-
First, import
import numpy as np import matplotlib.pyplot as plt plt.rcParams["font.family"] = "MS Gothic" -
Graph Basics
# Securing the drawing area fig = plt.figure() # Securing axes # If there is only one graph, 1,1,1 is fine. # This number changes when multiple graphs are arranged side by side, but this will be explained later. ax = fig.add_subplot(1, 1, 1) # Draw a line graph on the axis # Line graphs are plot, and scatter plots (without lines) are scatter # However, you can also draw scatter plots using plot ax.plot(np.array([1, 4, 7]), np.array([2, 5, 1])) # Line graph connecting (1,2),(4,5), and (7,1) # graph drawing plt.show()
-
mark with a dot (point)
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) # You can draw points at coordinates specified with the marker option # marker = "o" : round dot # marker = "x" : cross mark # marker = "^" : triangles ax.plot(np.array([1, 4, 7]), np.array([2, 5, 1]), marker = "o") plt.show()
-
Change line type
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # Line style can be changed with the linestyle option # linestyle = "dashed" # linestyle = "dotted" # linestyle = "dashdot" # linestyle = "none" : No line (can be used with markers to create a scatter plot) ax.plot(x, np.sin(x), linestyle = "dashed") plt.show()
-
Change the color
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # You can change the color using the color option (the color of the dots will also change at the same time) # color = "red" # color = "tab:red" # color = "black" ax.plot(x, np.sin(x), color = "tab:green") plt.show()
-
Name the axis
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # ax.set_xlabel assigns a name to the x-axis ax.set_xlabel("name of x-axis") # ax.set_ylabel assigns a name to the y-axis ax.set_ylabel("name of y-axis") ax.plot(x, np.sin(x)) plt.show()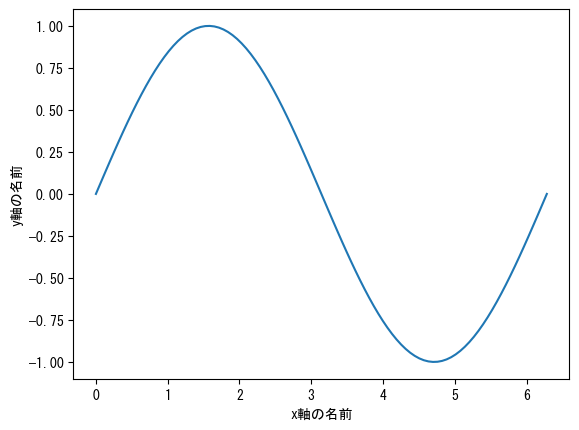
-
Name the graph
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # ax.set_title gives the graph a name ax.set_title("graph of sin(x)") ax.plot(x, np.sin(x)) plt.show()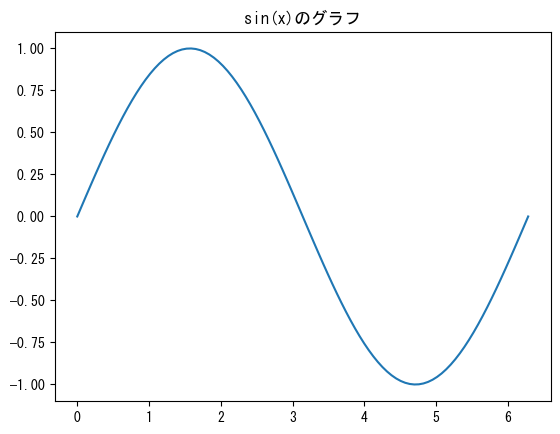
-
Set the axis range
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # You can change the range of the x-axis with ax.set_xlim ax.set_xlim(0, pi) # You can change the range of the y-axis with ax.set_ylim ax.set_ylim(-5, 5) ax.plot(x, np.tan(x)) plt.show()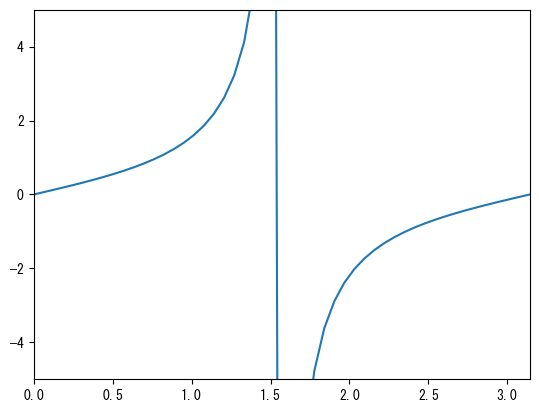
-
Set axis values
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # ax.set_xticks (vector) allows you to display only the desired parts of the x-axis values ax.set_xticks(np.array([0, np.pi/2, np.pi, 3*np.pi/2, 2*np.pi])) # ax.set_yticks (vector) allows you to display only the desired parts of the y-axis values ax.set_yticks(np.array([-1, -0.5, 0, 0.5, 1])) ax.plot(x, np.sin(x)) plt.show()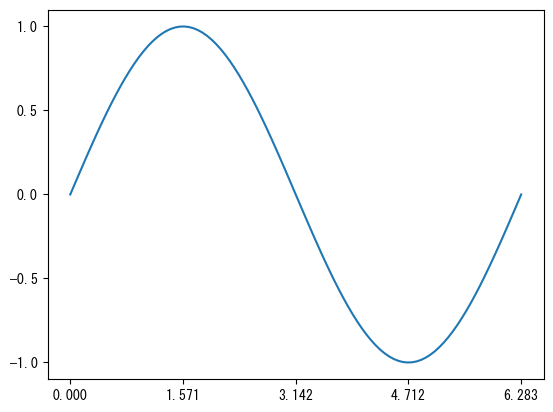
-
Set the values of the axes to whatever notation you want
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) ax.set_xticks(np.array([0, np.pi/2, np.pi, 3*np.pi/2, 2*np.pi])) # ax.set_xticklabels (vector) allows you to change the display of the values set with xticks # The number of elements in the vectors ax.set_xticks and ax.set_xtickslabels must be the same ax.set_xticklabels(np.array(["0", "π/2", "π", "3π/2", "2π"])) ax.set_yticks(np.array([-1, 0, 1])) # ax.set_yticklabels (vector) allows you to change the display of the values set with yticks # The number of elements in the vectors ax.set_yticks and ax.set_ytickslabels must be the same ax.set_yticklabels(np.array(["ymin", "0", "ymax"])) ax.plot(x, np.sin(x)) plt.show()
-
Grid against the scale
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) ax.set_xticks(np.array([0, np.pi/2, np.pi, 3*np.pi/2, 2*np.pi])) ax.set_xticklabels(np.array(["0", "π/2", "π", "3π/2", "2π"])) # ax.set_grid (axis = "x") adds a grid to the x-axis tick marks ax.grid(axis = "x") ax.set_yticks(np.array([-1, 0, 1])) ax.set_yticklabels(np.array(["ymin", "0", "ymax"])) # ax.set_grid (axis = "y") adds a grid to the y-axis tick marks ax.grid(axis = "y", color="tab:red", linestyle = "dashed") ax.plot(x, np.sin(x)) plt.show()
-
Attach a small scale with no numerical value
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) ax.set_xticks(np.array([0, np.pi/2, np.pi, 3*np.pi/2, 2*np.pi])) ax.set_xticklabels(np.array(["0", "π/2", "π", "3π/2", "2π"])) # minor = true option sets the minor tick mark setting ax.set_xticks(np.array([np.pi/4, 3*np.pi/4, 5*np.pi/4, 7*np.pi/4]), minor = True) ax.set_yticks(np.array([-1, 0, 1])) ax.set_yticklabels(np.array(["ymin", "0", "ymax"])) ax.plot(x, np.sin(x)) plt.show()
-
Attach grid to small scale
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) ax.set_xticks(np.array([0, np.pi/2, np.pi, 3*np.pi/2, 2*np.pi])) ax.set_xticklabels(np.array(["0", "π/2", "π", "3π/2", "2π"])) ax.set_xticks(np.array([np.pi/4, 3*np.pi/4, 5*np.pi/4, 7*np.pi/4]), minor = True) ax.grid(axis = "x") # which = "minor" option makes the grid for minor tick marks ax.grid(axis = "x", which = "minor", linestyle = "dashed") ax.set_yticks(np.array([-1, 0, 1])) ax.set_yticklabels(np.array(["ymin", "0", "ymax"])) ax.plot(x, np.sin(x)) plt.show()
-
Same ratio of length to number on x-axis and y-axis
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # ax.set_aspect(1) sets the ratio of the vertical and horizontal values to be the same ax.set_aspect(1) ax.plot(x, np.sin(x)) plt.show()
-
Using LaTeX for Strings
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # By adding an L before a string, you can use LaTeX notation # ax.set_x(y)label, ax.set_title, ax.set_x(y)ticklabels, label (described later) etc. correspond to this ax.set_xlabel(r"$x$") ax.set_ylabel(r"$\sin(x)$") ax.set_title(r"graph of $\sin(x)$") ax.set_xticks(np.array([0, np.pi/2, np.pi, 3*np.pi/2, 2*np.pi])) ax.set_xticklabels(np.array(["0", r"$\pi/2$", r"$\pi$", r"$3\pi/2$", r"$2\pi$"])) ax.set_yticks(np.array([-1, 0, 1])) ax.set_yticklabels(np.array([r"$y_\mathrm{min}$", "0", r"$y_\mathrm{max}$"])) ax.plot(x, np.sin(x)) plt.show()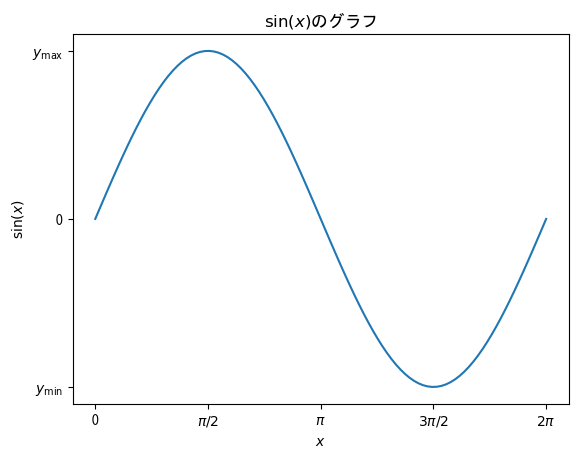
-
Graph size
# You can change the size of the drawing area with the # figsize option # The default is [6.4; 4.8] fig = plt.figure(figsize = np.array([6.4, 4.8]) * 0.75) ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) ax.plot(x, np.sin(x)) plt.show()
-
Saving Graphs
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) ax.plot(x, np.sin(x)) # Instead of show(), use savefig("filename") to save the graph # By setting bbox_inches = "tight" and pad_inches = 0.1 as options, you can minimize the extra space around the image # Commonly used file extensions include png and pdf (the file type is automatically determined based on the file extension) plt.savefig("graph.png", bbox_inches = "tight", pad_inches = 0.1)
-
Draw multiple graphs on a single axis
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # By repeatedly using ax.plot and ax.scatter, you can draw multiple graphs on a single axis # The color changes automatically (of course, you can specify each color individually with the color option) ax.plot(x, np.sin(x)) ax.plot(x, np.cos(x)) plt.show()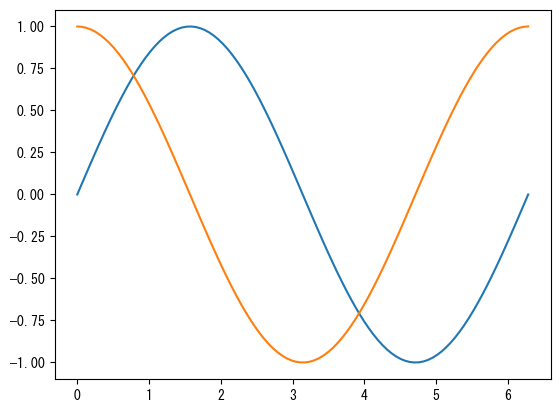
-
Mixed plot and scatter
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x1 = np.linspace(0, 2*np.pi, 100) x2 = np.linspace(0, 2*np.pi, 10) # You can also mix ax.plot and ax.scatter # Note that color management is separate for plot and scatter ax.plot(x1, np.sin(x1)) ax.scatter(x2, np.cos(x2)) plt.show()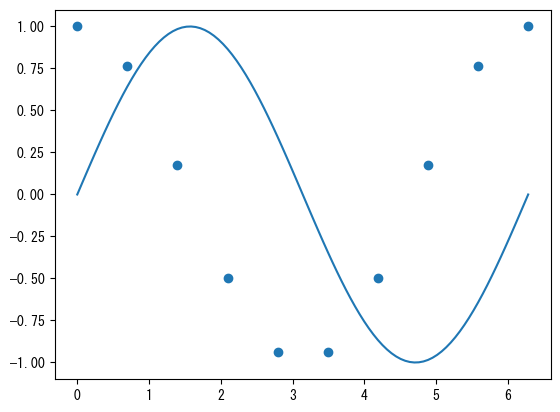
-
Show legend
fig = plt.figure() ax = fig.add_subplot(1, 1, 1) x = np.linspace(0, 2*np.pi, 100) # Name with the label option ax.plot(x, np.sin(x), label = r"$\sin(x)$") ax.plot(x, np.cos(x), label = r"$\cos(x)$") # With ax.legend(), you can display the names assigned with the label option as a legend ax.legend() plt.show()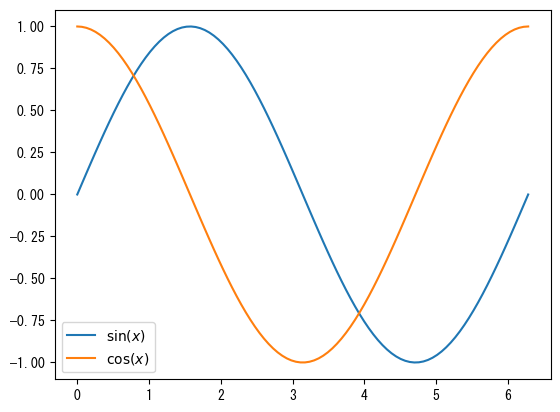
-
Legend Location
fig = plt.figure() ax = fig.add_subplot(1,1,1) x = np.linspace(0, 2*np.pi, 100) ax.plot(x, np.sin(x), label = r"$\sin(x)$") ax.plot(x, np.sin(x - 2*np.pi/5), label = r"$\sin(x-2\pi/5)$") ax.plot(x, np.sin(x - 4*np.pi/5), label = r"$\sin(x-4\pi/5)$") ax.plot(x, np.sin(x - 6*np.pi/5), label = r"$\sin(x-6\pi/5)$") ax.plot(x, np.sin(x - 8*np.pi/5), label = r"$\sin(x-8\pi/5)$") # The position of the legend is adjusted automatically, but you can specify the approximate position using loc # "upper left" # "upper center" # "upper right" # "center left" # "center" # "center right" # "lower left" # "lower center" # "lower right" ax.legend(loc = "center") plt.show()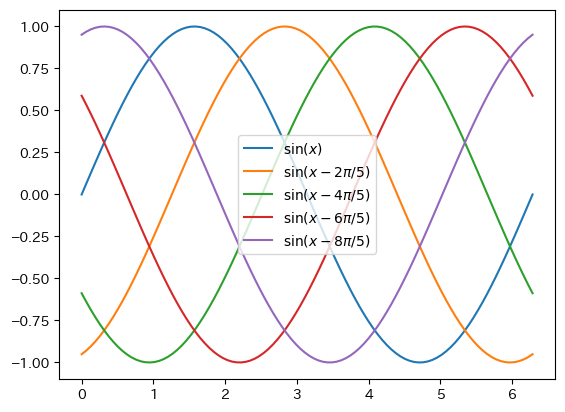
-
Detailed location of legend
fig = plt.figure() ax = fig.add_subplot(1,1,1) x = np.linspace(0, 2*np.pi, 100) ax.plot(x, np.sin(x), label = r"$\sin(x)$") ax.plot(x, np.sin(x - 2*np.pi/5), label = r"$\sin(x-2\pi/5)$") ax.plot(x, np.sin(x - 4*np.pi/5), label = r"$\sin(x-4\pi/5)$") ax.plot(x, np.sin(x - 6*np.pi/5), label = r"$\sin(x-6\pi/5)$") ax.plot(x, np.sin(x - 8*np.pi/5), label = r"$\sin(x-8\pi/5)$") # You can specify the position with bbox_to_anchor. (0,0) is the lower left corner, (1,1) is the upper right corner # Furthermore, by specifying loc, you can set the position of the legend to bbox_to_anchor # Specify the number of columns with ncols ax.legend(bbox_to_anchor = (0.5, 1), loc = "lower center", ncols = 5) plt.show()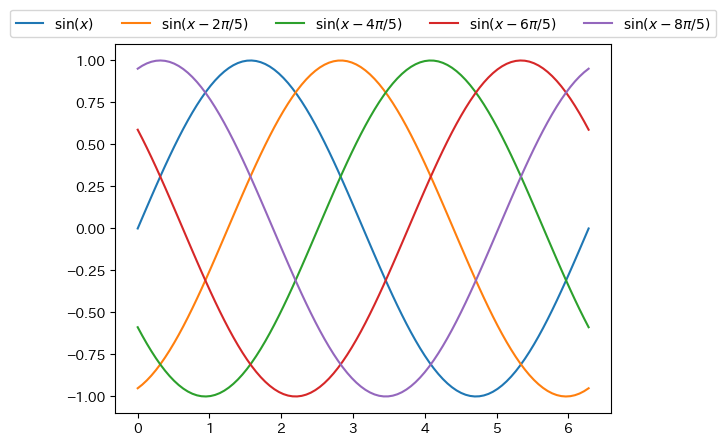
-
Different y-axis for left and right
fig = plt.figure() t = np.linspace(0, 0.9, 100) # First, create a graph for the left side (normal) of the y-axis ax1 = fig.add_subplot(1, 1, 1) ax1.plot(t, 5 - 9.8 * t) ax1.set_xlabel("time [s]") ax1.set_ylabel("velocity [m/s]") # Create a graph for the y-axis on the right side # Creating an axis with the y-axis on the right using twinx ax2 = ax1.twinx() # Please note that color management is separate for ax1 and ax2 ax2.plot(t, - 4.9 * t ** 2 + 5 * t) ax2.set_ylabel("height [m]") plt.show()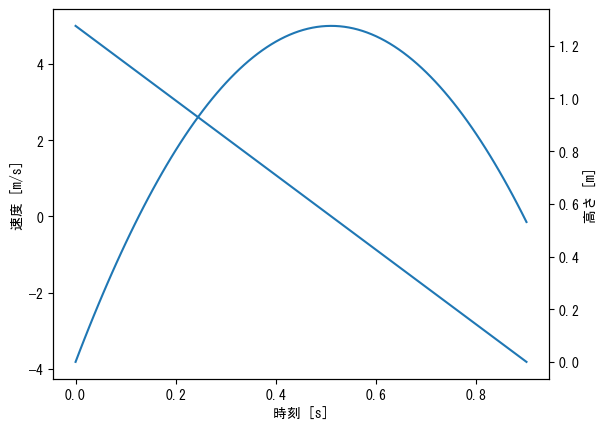
-
Different y-axis for left and right (color, legend adjustment)
fig = plt.figure() t = np.linspace(0, 0.9, 100) ax1 = fig.add_subplot(1, 1, 1) ax1.plot(t, 5 - 9.8 * t, label = "velocity (without friction) ") ax1.plot(t, 24.6 * np.exp(- t / 2) - 19.6, label = "velocity (with friction) ") ax1.set_xlabel("time [s]") ax1.set_ylabel("velocity [m/s]") ax1.legend() ax2 = ax1.twinx() ax2.plot(t, - 4.9 * t ** 2 + 5 * t, label = "height (without friction) ", color = "tab:green") ax2.plot(t, 49.2 * (1 - np.exp(- t / 2)) - 19.6 * t, label = "height (with friction) ", color = "tab:red") ax2.set_ylabel("height [m]") ax2.legend() plt.show()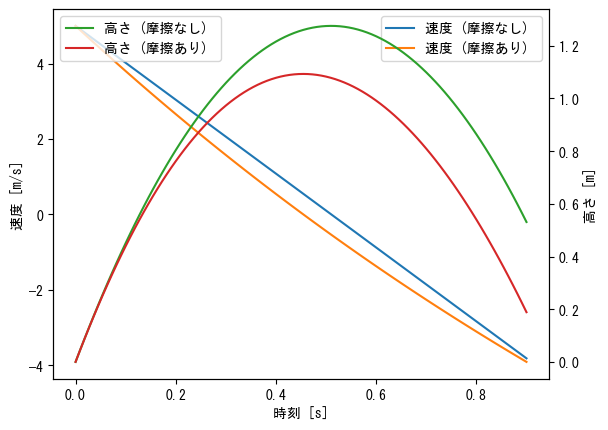
-
Multiple axes
# Be sure to set the size of the drawing area, as multiple axes will be drawn fig = plt.figure(figsize = np.array([6.4 * 2, 4.8 * 3]) * 0.8) x1 = np.linspace(- np.pi, np.pi, 100) x2 = np.linspace(-1, 1, 100) x3 = np.linspace(-2, 2, 100) x4 = np.linspace(1, 2, 100) # The first one when the axes are arranged in three rows and two columns ax = fig.add_subplot(3, 2, 1) ax.plot(x1, np.sin(x1), label = r"$\sin(x)$") ax.plot(x1, np.cos(x1), label = r"$\cos(x)$") ax.plot(x1, np.tan(x1), label = r"$\tan(x)$") ax.set_ylim(-1.5, 1.5) ax.set_xlabel(r"$x$") ax.set_title("Graph of trigonometric functions") ax.legend() # The second one when the axes are arranged in three rows and two columns ax = fig.add_subplot(3, 2, 2) ax.plot(x3, np.sinh(x3), label = r"$\sinh(x)$") ax.plot(x3, np.cosh(x3), label = r"$\cosh(x)$") ax.plot(x3, np.tanh(x3), label = r"$\tanh(x)$") ax.set_xlabel(r"$x$") ax.set_title("Graph of hyperbolic functions") ax.legend() # The third one when the axes are arranged in three rows and two columns ax = fig.add_subplot(3, 2, 3) ax.plot(x1, 1 / np.sin(x1), label = r"$\csc(x)$") ax.plot(x1, 1 / np.cos(x1), label = r"$\sec(x)$") ax.plot(x1, 1 / np.tan(x1), label = r"$\cot(x)$") ax.set_ylim(-10, 10) ax.set_xlabel(r"$x$") ax.set_title("Graph of the reciprocal of trigonometric functions") ax.legend() # The fourth one when the axes are arranged in three rows and two columns ax = fig.add_subplot(3, 2, 4) ax.plot(x3, 1 / np.sinh(x3), label = r"$\mathrm{csch}(x)$") ax.plot(x3, 1 / np.cosh(x3), label = r"$\mathrm{sech}(x)$") ax.plot(x3, 1 / np.tanh(x3), label = r"$\coth(x)$") ax.set_ylim(-4, 4) ax.set_xlabel(r"$x$") ax.set_title("Graph of the reciprocal of hyperbolic functions") ax.legend() # The fifth one when the axes are arranged in three rows and two columns ax = fig.add_subplot(3, 2, 5) ax.plot(x2, np.arcsin(x2), label = r"$\sin^{-1}(x)$") ax.plot(x2, np.arccos(x2), label = r"$\cos^{-1}(x)$") ax.plot(x3, np.arctan(x3), label = r"$\tan^{-1}(x)$") ax.set_xlabel(r"$x$") ax.set_title("Graph of inverse trigonometric functions") ax.legend() # The sixth one when the axes are arranged in three rows and two columns ax = fig.add_subplot(3, 2, 6) ax.plot(x3, np.arcsinh(x3), label = r"$\sinh^{-1}(x)$") ax.plot(x4, np.arccosh(x4), label = r"$\cosh^{-1}(x)$") ax.plot(x2, np.arctanh(x2), label = r"$\tanh^{-1}(x)$") ax.set_xlabel(r"$x$") ax.set_title("Graph of inverse hyperbolic functions") ax.legend() # Adjust axis spacing with subplots_adjust # wspace for horizontal, hspace for vertical plt.subplots_adjust(wspace=0.2, hspace=0.3) plt.show()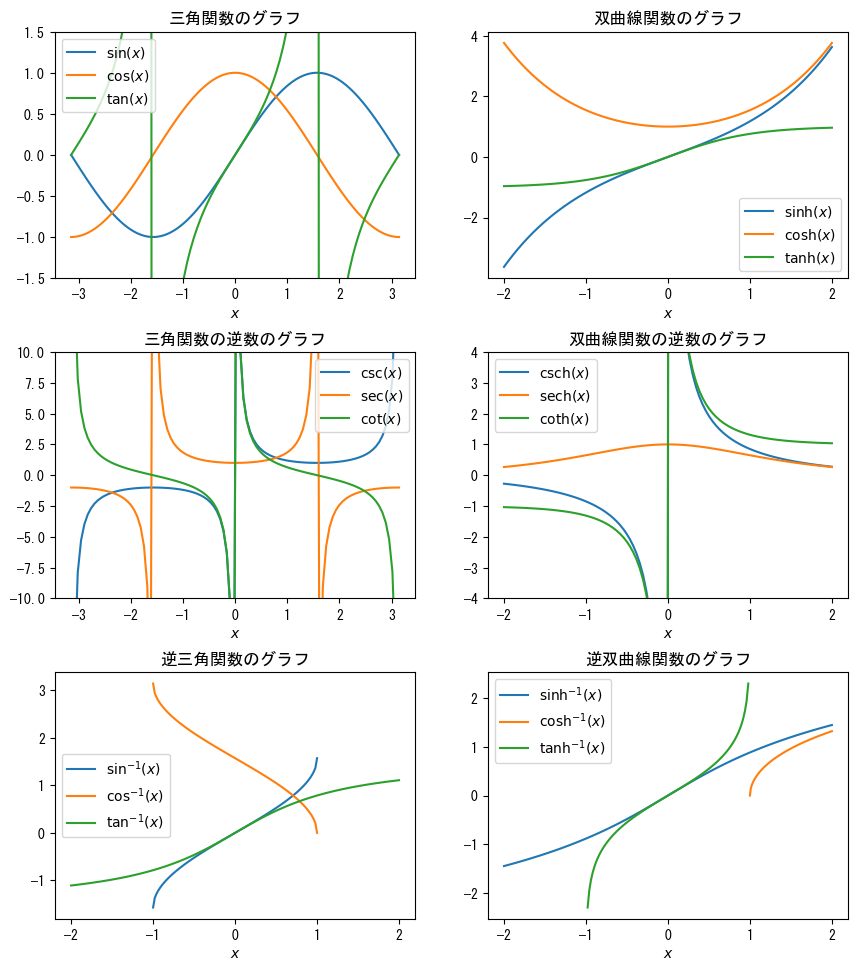
-
single-logarichmic graph
fig = plt.figure() x = np.linspace(0, 5, 5) ax = fig.add_subplot(1, 1, 1) # ax.set_yscale("log") creates a semi-logarithmic graph (y-axis is logarithmic scale) ax.set_yscale("log") ax.plot(x, 5 * np.exp(- 2 * x), marker = "o") ax.set_title("In a semi-logarithmic graph, exponential functions appear as straight lines") ax.set_xlabel(r"$x$") ax.set_ylabel(r"$5e^{-2x}$") plt.show()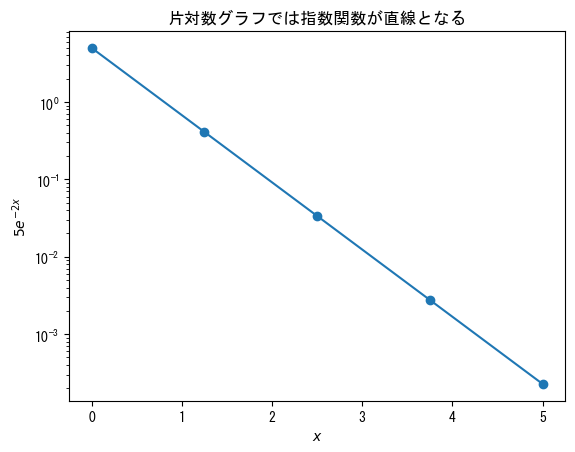
-
double-logarithmic graph
fig = plt.figure() x = np.linspace(1, 100, 10) ax = fig.add_subplot(1, 1, 1) # Add ax.set_xscale("log") to create a double logarithmic graph (logarithmic scale on both the x-axis and y-axis) ax.set_xscale("log") ax.set_yscale("log") ax.plot(x, 10 * x ** (- 5 / 3), marker = "o") ax.set_title("In a logarithmic graph, power functions become straight lines") ax.set_xlabel(r"$x$") ax.set_ylabel(r"$10x^{-5/3}$") plt.show()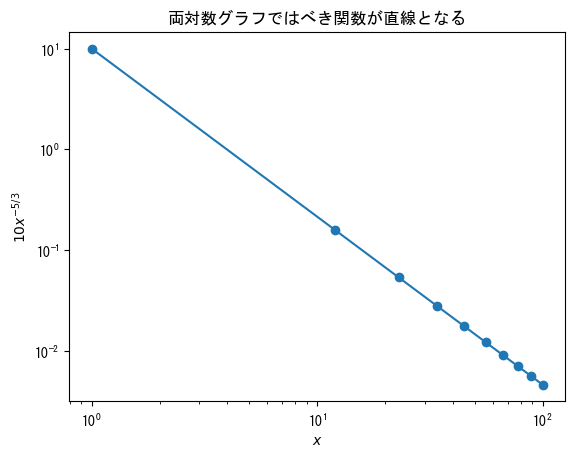
-
When using ticks (setting axis values) in a logarithmic graph, it is best to use them in conjunction with ticklabels (or else the graph may not display well).
# On logarithmic scale graphs, minor ticks are basically displayed as they are # If you want to delete it, add the following description plt.rcParams["ytick.minor.size"] = 0 # Set the vector of values you want to set on the y-axis fig = plt.figure() x = np.linspace(0, 5, 5) ax = fig.add_subplot(1, 1, 1) ax.set_yscale("log") # Set the vector of values you want to set on the y-axis vector_y = np.array([0.02, 0.7, 1.4, 5]) ax.set_yticks(vector_y) ax.set_yticklabels(vector_y) ax.plot(x, 5 * np.exp(- 2 * x), marker = "o") ax.set_xlabel(r"$x$") ax.set_ylabel(r"$5e^{-2x}$") plt.show()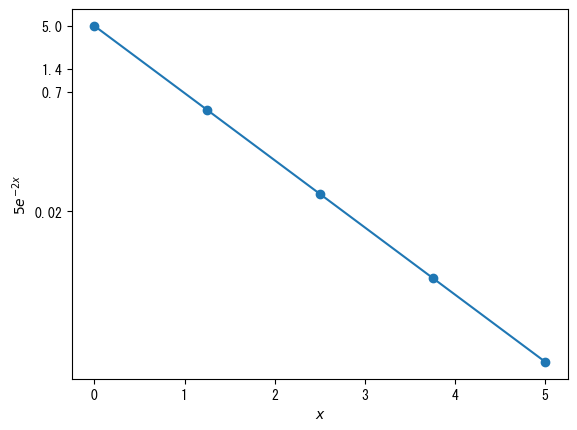
-
Basic Histogram
fig = plt.figure() x = np.hstack([np.random.normal(0, 1, 1000), np.random.normal(5, 2, 1000)]) ax = fig.add_subplot(1, 1, 1) # ax.hist(vector) produces a histogram ax.hist(x) ax.set_xlabel(r"$x$") ax.set_ylabel("histogram") plt.show()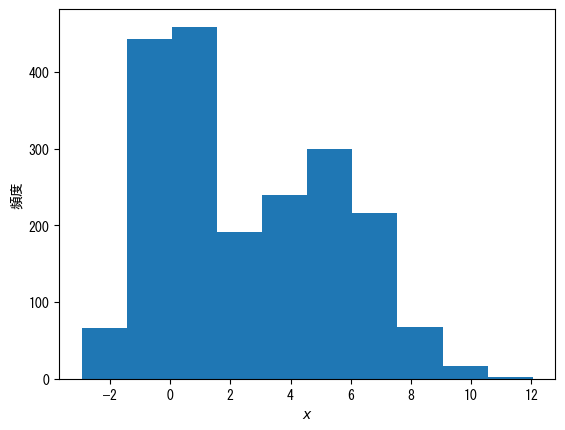
-
change the grade number
fig = plt.figure() x = np.hstack([np.random.normal(0, 1, 1000), np.random.normal(5, 2, 1000)]) ax = fig.add_subplot(1, 1, 1) # Specify the number of classes with the bins option ax.hist(x, bins = 20) ax.set_xlabel(r"$x$") ax.set_ylabel("histogram") plt.show()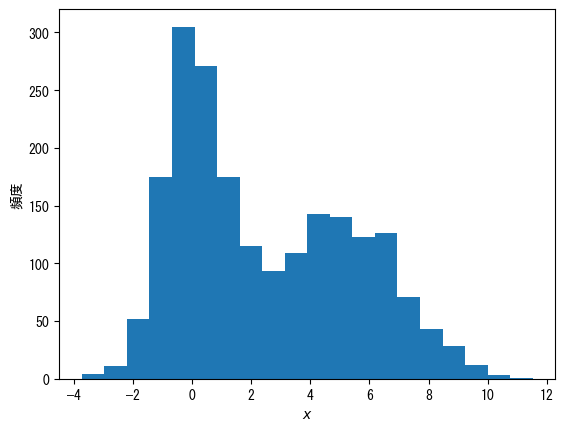
-
Make it a probability distribution, not a frequency
fig = plt.figure() x = np.hstack([np.random.normal(0, 1, 1000), np.random.normal(5, 2, 1000)]) ax = fig.add_subplot(1, 1, 1) # When density = true, it becomes a probability distribution rather than a frequency distribution ax.hist(x, bins = 20, density = True) ax.set_xlabel(r"$x$") ax.set_ylabel(r"$p(x)$") plt.show()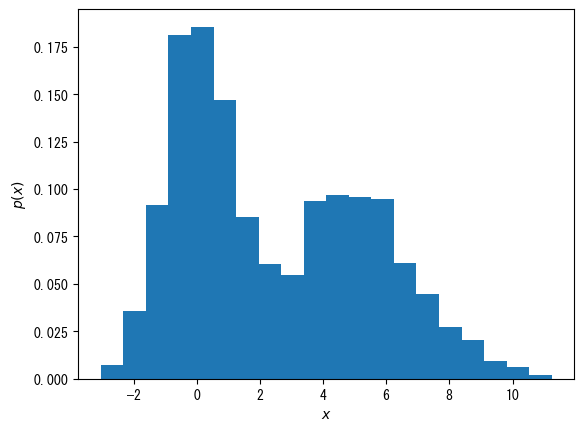
-
Bars only on the outer border (useful for displaying multiple histograms)
fig = plt.figure() x1 = np.random.normal(0, 1, 1000) x2 = np.random.normal(5, 2, 1000) x3 = np.hstack([x1, x2]) ax = fig.add_subplot(1, 1, 1) # When histtype = "step", only the outer frame is displayed ax.hist(x1, bins = 20, density = True, histtype = "step", label = r"$p_1(x)$") ax.hist(x2, bins = 20, density = True, histtype = "step", label = r"$p_2(x)$") ax.hist(x3, bins = 20, density = True, histtype = "step", label = r"$p_{1 + 2}(x)$") ax.set_xlabel(r"$x$") ax.set_ylabel(r"$p(x)$") ax.legend() plt.show()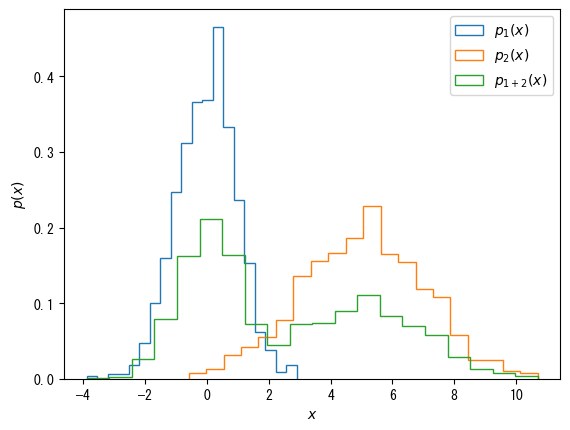
-
A note on histograms
If you want to plot something more advanced, such as a histogram of a line chart, you have to get a vector of the histogram, which can be obtained by the fit function in StatsBase, but it is quite tedious (this is where Python ( numpy) wins hands down).
Point color and point size in scatter plots by scatter
-
Specify a different vector for the color of a point in a scatterplot with scatter
fig = plt.figure() height = np.array([172.3, 165, 179.6, 174.5, 173.8, 165.4, 164.5, 174.9, 166.8, 185]) mass = np.array([75.24, 55.8, 78, 71.1, 67.7, 55.4, 63.7, 77.2, 67.5, 84.6]) fat = np.array([21.3, 15.7, 20.1, 18.4, 17.1, 22, 32.2, 36.9, 27.6, 14.4]) ax = fig.add_subplot(1, 1, 1) # By specifying a vector with the c option, the points will be colored according to the values in that vector ax.scatter(height, mass, c = fat) ax.set_xlabel("height [cm]") ax.set_ylabel("weight [kg]") plt.show()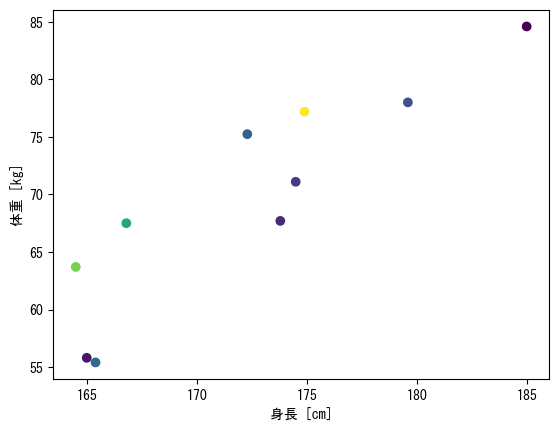
-
Display a color bar that maps colors to numerical values
fig = plt.figure() height = np.array([172.3, 165, 179.6, 174.5, 173.8, 165.4, 164.5, 174.9, 166.8, 185]) mass = np.array([75.24, 55.8, 78, 71.1, 67.7, 55.4, 63.7, 77.2, 67.5, 84.6]) fat = np.array([21.3, 15.7, 20.1, 18.4, 17.1, 22, 32.2, 36.9, 27.6, 14.4]) ax = fig.add_subplot(1, 1, 1) # Assign the scatter plot itself as a variable (in this case, the variable c_points) c_points = ax.scatter(height, mass, c = fat) # colorbar(c_points, ax = ax) adds a color bar fig.colorbar(c_points) ax.set_xlabel("height [cm]") ax.set_ylabel("weight [kg]") plt.show()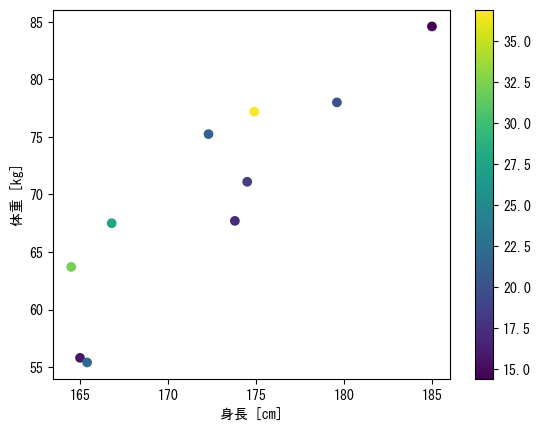
-
Option 1 for color, color bar
fig = plt.figure() height = np.array([172.3, 165, 179.6, 174.5, 173.8, 165.4, 164.5, 174.9, 166.8, 185]) mass = np.array([75.24, 55.8, 78, 71.1, 67.7, 55.4, 63.7, 77.2, 67.5, 84.6]) fat = np.array([21.3, 15.7, 20.1, 18.4, 17.1, 22, 32.2, 36.9, 27.6, 14.4]) ax = fig.add_subplot(1, 1, 1) # The vmax and vmin options allow you to set the maximum and minimum values for colors c_points = ax.scatter(height, mass, c = fat, vmin = 10, vmax = 40) # Color bars are named with the label option fig.colorbar(c_points, label = "body fat [%]") ax.set_xlabel("height [cm]") ax.set_ylabel("weight [kg]") plt.show()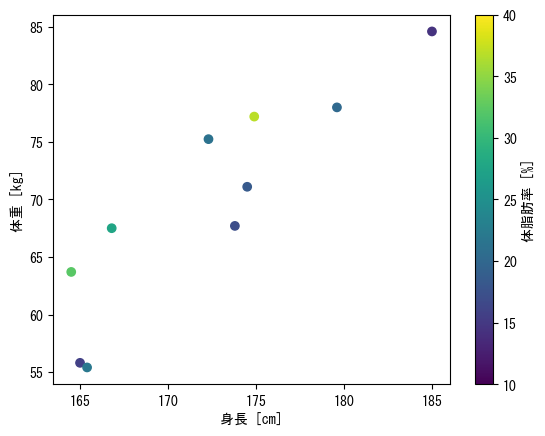
-
Option 2 for color, color bar
fig = plt.figure() height = np.array([172.3, 165, 179.6, 174.5, 173.8, 165.4, 164.5, 174.9, 166.8, 185]) mass = np.array([75.24, 55.8, 78, 71.1, 67.7, 55.4, 63.7, 77.2, 67.5, 84.6]) fat = np.array([21.3, 15.7, 20.1, 18.4, 17.1, 22, 32.2, 36.9, 27.6, 14.4]) ax = fig.add_subplot(1, 1, 1) # Colors can be changed with the cmap option # viridis : Default blue→yellow # jet : rainbow colors # hsv : periodic rainbow colors c_points = ax.scatter(height, mass, c = fat, cmap = "hsv") # The value set in the ticks option is displayed # Furthermore, assign the color bar itself to another variable (in this case, cbar), and use cbar.ax.set_yticklabels to change the display cbar = fig.colorbar(c_points, label = "body fat [%]", ticks = np.array([15, 25, 35])) cbar.ax.set_yticklabels(np.array(["low", "mid", "high"])) ax.set_xlabel("height [cm]") ax.set_ylabel("weight [kg]") plt.show()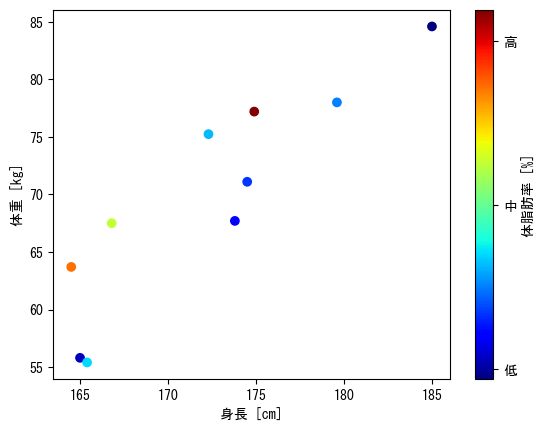
-
Dot size
fig = plt.figure() height = np.array([172.3, 165, 179.6, 174.5, 173.8, 165.4, 164.5, 174.9, 166.8, 185]) mass = np.array([75.24, 55.8, 78, 71.1, 67.7, 55.4, 63.7, 77.2, 67.5, 84.6]) fat = np.array([21.3, 15.7, 20.1, 18.4, 17.1, 22, 32.2, 36.9, 27.6, 14.4]) age = np.array([27, 25, 31, 32, 28, 36, 42, 33, 54, 28]) ax = fig.add_subplot(1, 1, 1) # You can choose the size of the dots as an option # (markersize for plots) # However, in the case of scatter, you can change the size of each point by specifying a vector in c c_points = ax.scatter(height, mass, c = fat, s = age * 4) cbar = fig.colorbar(c_points, label = "body fat [%]", ticks = np.array([15, 25, 35])) cbar.ax.set_yticklabels(np.array(["low", "mid", "high"])) ax.set_xlabel("height [cm]") ax.set_ylabel("weight [kg]") ax.set_title("Marker size corresponds to age") plt.show()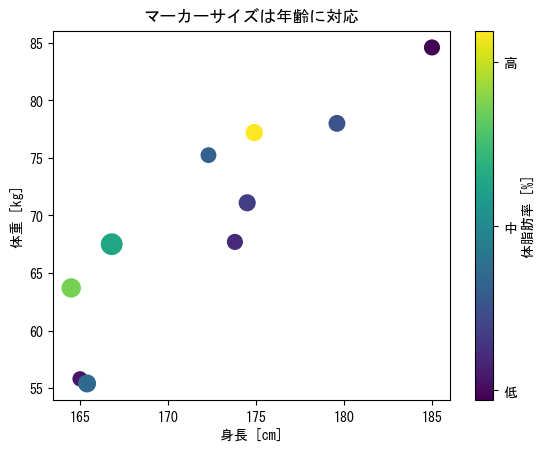
-
imshow is like displaying a matrix as you see it
fig = plt.figure() matrix = np.array([[2, 3, 0, 0], [1, 2, 3, 0], [0, 1, 2, 3]]) ax = fig.add_subplot(1, 1, 1) # min, vmax, and cmap options can also be used (see scatter) c_matrix = ax.imshow(matrix) # Color bars can also be used # label and ticks options are also available fig.colorbar(c_matrix, label = "matrix value") ax.set_xlabel("column") ax.set_ylabel("row") plt.show()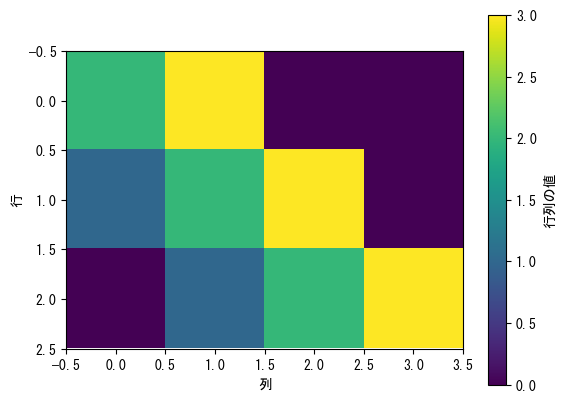
-
If the height of the color bar is uncomfortable
fig = plt.figure() matrix = np.array([[2, 3, 0, 0], [1, 2, 3, 0], [0, 1, 2, 3]]) ax = fig.add_subplot(1,1,1) c_matrix = ax.imshow(matrix) # The quickest way is to set fraction = (row size) / (column size) * 0.046 and pad = 0.04. fig.colorbar(c_matrix, label = "matrix value", fraction = 3 / 4 * 0.046, pad = 0.04) ax.set_xlabel("column") ax.set_ylabel("row") plt.show()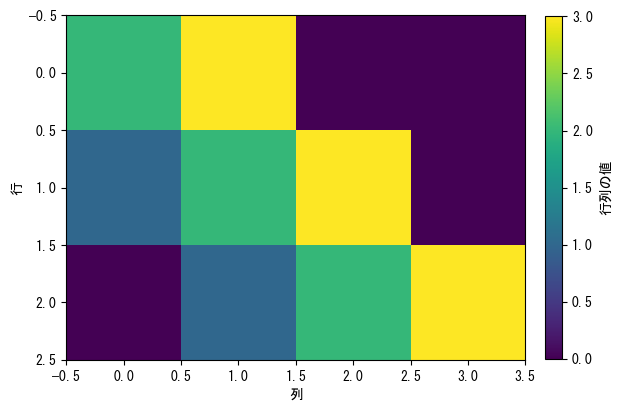
-
pcolor is an image like contour lines, where x and y coordinates can be specified
However, if nothing is specified, the line direction of the matrix will be x and the columns will be y direction (opposite to imshow)fig = plt.figure() # Create a vector for the values of x and y x = np.linspace(-2*np.pi, 2*np.pi, 100) y = np.linspace(-np.pi, np.pi, 100) # Convert x and y into a matrix x, y = np.meshgrid(x, y) ax = fig.add_subplot(1, 1, 1) # The basic format is ax.pcolor(matrix specifying x coordinates, matrix specifying y coordinates, matrix for heat map, shading = "auto") # The vmin, vmax, and cmap options can also be used (see scatter) c_pcolor = ax.pcolor(x, y, np.sin(x + y), shading = "auto") # Color bars can also be used # label and ticks options are also available fig.colorbar(c_pcolor, label = r"$\sin(x+y)$") ax.set_xlabel(r"$x$") ax.set_ylabel(r"$y$") ax.set_xticks(np.array([-2*np.pi, -np.pi, 0, np.pi, 2*np.pi])) ax.set_xticklabels(np.array([r"$-2\pi$", r"$-\pi$", r"$0$", r"$\pi$", r"$2\pi$"])) ax.set_yticks(np.array([-np.pi, 0, np.pi])) ax.set_yticklabels(np.array([r"$-\pi$", r"$0$", r"$\pi$"])) plt.show()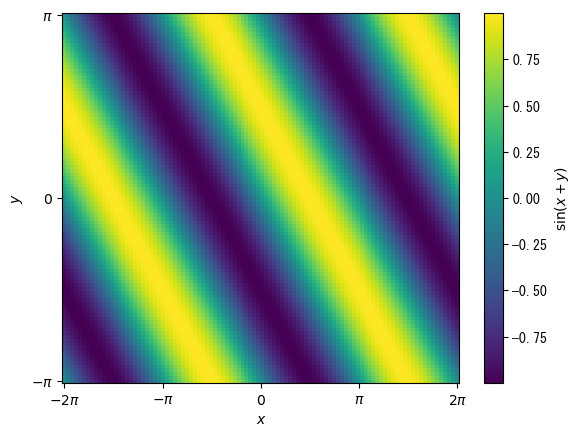
-
Basics of 3D Graphs
# 3D graphs are basically convenient because you can freely change the viewpoint using the mouse # To make this possible, it is necessary to draw the graph in a separate window # If you set %matplotlib tk (or qt) , the graph will open in a separate window # Conversely, if you set %matplotlib inline, the graph will be displayed on the Jupyter notebook %matplotlib tk fig = plt.figure() t = np.linspace(0, 2*np.pi, 100) x = np.sin(t) y = np.cos(t) z = t + np.sin(2 * t) # Adding "3d" to projection makes it a 3D graph ax = fig.add_subplot(1, 1, 1, projection = "3d") # A plot is a 3D line graph that also requires a vector for the z-coordinate ax.plot(x, y, z) # ax.set_zlabel, ax.set_zticks, ax.set_zticklabels, and ax.set_zlim can also be used ax.set_xlabel(r"$x = \sin(t)$") ax.set_ylabel(r"$y = \cos(t)$") ax.set_zlabel(r"$z = t + \sin(2t)$") plt.show()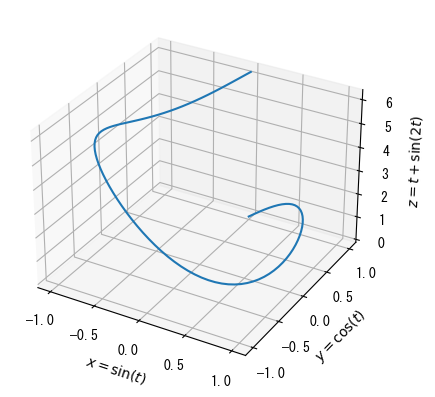
-
Dispersal Chart
%matplotlib tk fig = plt.figure() age = np.array([27, 25, 31, 32, 28, 36, 42, 33, 54, 28]) height = np.array([172.3, 165, 179.6, 174.5, 173.8, 165.4, 164.5, 174.9, 166.8, 185]) mass = np.array([75.24, 55.8, 78, 71.1, 67.7, 55.4, 63.7, 77.2, 67.5, 84.6]) fat = np.array([21.3, 15.7, 20.1, 18.4, 17.1, 22, 32.2, 36.9, 27.6, 14.4]) ax = fig.add_subplot(1, 1, 1, projection = "3d") # 3D scatter can specify vectors corresponding to colors in the same way as 2D scatter c_points = ax.scatter(height, mass, fat, c = age) # The color bars are close to the graph when set to default, so it is better to move them away a little using the pad option plt.colorbar(c_points, label = "年齢 [歳]", pad = 0.15) ax.set_xlabel("height [cm]") ax.set_ylabel("weight [kg]") ax.set_zlabel("body fat [%]") plt.show()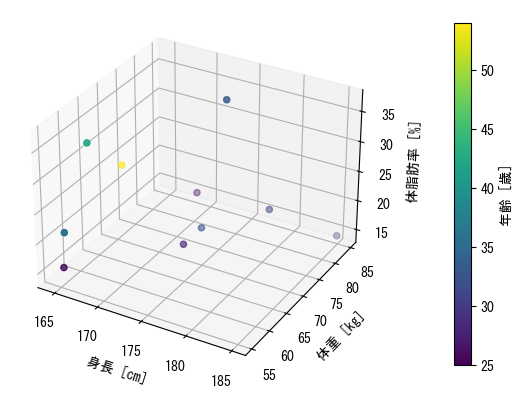
-
Curved surface (wireframe): alternative to pcolor
%matplotlib tk fig = plt.figure() x = np.linspace(- 2*np.pi, 2*np.pi,100) y = np.linspace(- np.pi, np.pi, 100) x, y = np.meshgrid(x, y) ax = fig.add_subplot(1, 1, 1, projection = "3d") # Displaying curved surfaces with ax.plot_wireframe ax.plot_wireframe(x, y, np.sin(x) + np.cos(y)) ax.set_xlabel(r"$x$") ax.set_ylabel(r"$y$") ax.set_zlabel(r"$\sin(x)+\cos(y)$") ax.set_xticks(np.array([-2*np.pi, -np.pi, 0, np.pi, 2*np.pi])) ax.set_xticklabels(np.array([r"$-2\pi$", r"$-\pi$", r"$0$", r"$\pi$", r"$2\pi$"])) ax.set_yticks(np.array([-np.pi, 0, np.pi])) ax.set_yticklabels(np.array([r"$-\pi$", r"$0$", r"$\pi$"])) plt.show()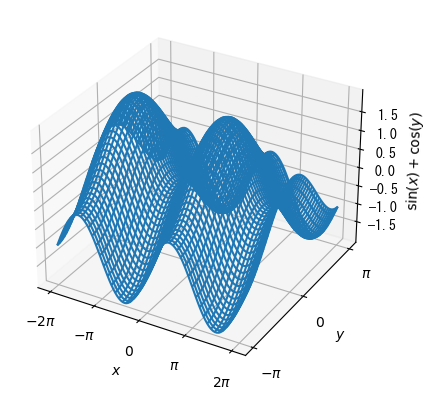
-
colored surface
%matplotlib tk fig = plt.figure() x = np.linspace(- 2*np.pi, 2*np.pi,100) y = np.linspace(- np.pi, np.pi, 100) x, y = np.meshgrid(x, y) ax = fig.add_subplot(1, 1, 1, projection = "3d") # Color-coded surface display with ax.plot_surface with cmap option c_surface = ax.plot_surface(x, y, np.sin(x) + np.cos(y), cmap = "viridis") plt.colorbar(c_surface, pad = 0.15) ax.set_xlabel(r"$x$") ax.set_ylabel(r"$y$") ax.set_zlabel(r"$\sin(x)+\cos(y)$") ax.set_xticks(np.array([-2*np.pi, -np.pi, 0, np.pi, 2*np.pi])) ax.set_xticklabels(np.array([r"$-2\pi$", r"$-\pi$", r"$0$", r"$\pi$", r"$2\pi$"])) ax.set_yticks(np.array([-np.pi, 0, np.pi])) ax.set_yticklabels(np.array([r"$-\pi$", r"$0$", r"$\pi$"])) plt.show()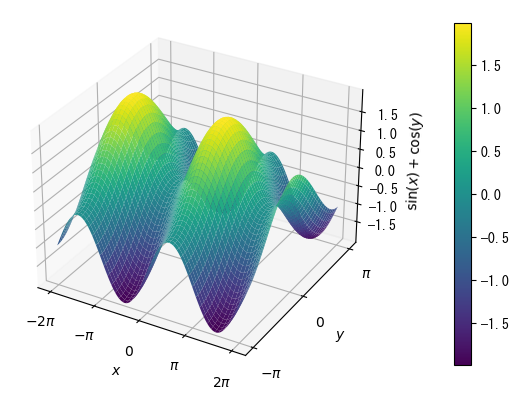
-
The animation is created in the manner of a so-called flipbook.
That is, the graph is displayed for a certain (short) amount of time, and the action is repeated.
In the case of PyPlot, the graph is repeatedly erased and then displayed.# Animations are displayed in a separate window, similar to 3D graphs # Incidentally, saving animations in PyPlot is troublesome, so it is better to give up on that %matplotlib tk fig = plt.figure() ax = fig.add_subplot(1, 1, 1) q = np.pi / 4 v0 = 5.0 v0x = v0 * np.cos(q) v0y = v0 * np.sin(q) g = 9.8 t = np.array([0.0]) x = np.array([0.0]) y = np.array([0.0]) # For loop to display animation for it in range(200): t_now = it * 0.0035 x_now = v0x * t_now y_now = v0y * t_now - 0.5 * g * t_now ** 2 t = np.append(t, t_now) x = np.append(x, x_now) y = np.append(y, y_now) # Graph drawing # Delete the previous graph ax.cla() # Plotting a new graph ax.scatter(x_now, y_now) ax.plot(x, y) ax.set_xlabel("horizontal distance [m]") ax.set_ylabel("vertical distance [m]") # It is better to decide the maximum and minimum values for the axis in advance ax.set_xlim(0, 3) ax.set_ylim(0, 0.8) ax.set_title("t = " + "{:5.3f}".format(t_now) + " [sec]") # Draw a graph and wait for the time (seconds) specified in () plt.pause(0.05)